Phylogenetic relationship between strains KKUY-0036, KKUY-0078
Por um escritor misterioso
Last updated 04 fevereiro 2025

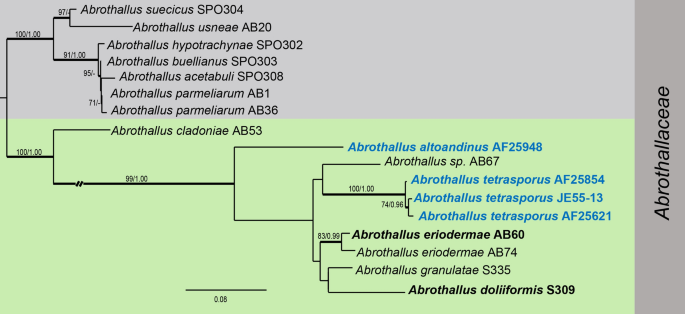
Fungal diversity notes 1611–1716: taxonomic and phylogenetic contributions on fungal genera and species emphasis in south China

Xantusiid “night” lizards: a puzzling phylogenetic problem revisited using likelihood-based Bayesian methods on mtDNA sequences - ScienceDirect

Saad ALAMRI, King Khalid University, Abha, KKU, College of Science

Phylogenetic tree depicting the relationship among Pseudomonas species.
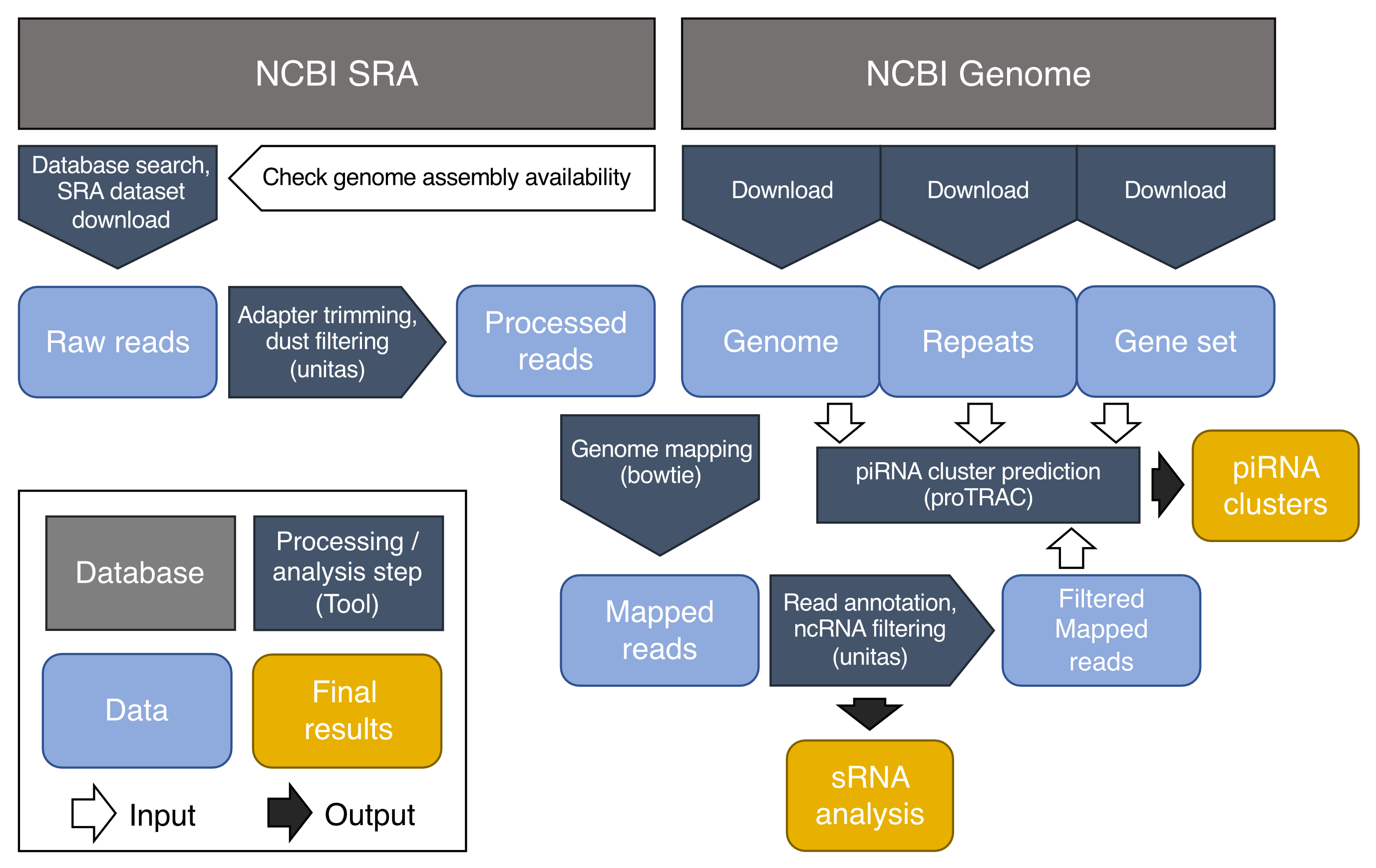
piRNA cluster database

WO2011046614A2 - Methods and systems for phylogenetic analysis - Google Patents

Phylogenetic relationship among the analysed strains. The phylogenetic

From Diagnosis to Treatment: Recent Advances in Patient-Friendly Biosensors and Implantable Devices

e Cell density (OD. 600 nm) time-courses during growth of the three

WO2011046614A2 - Methods and systems for phylogenetic analysis - Google Patents
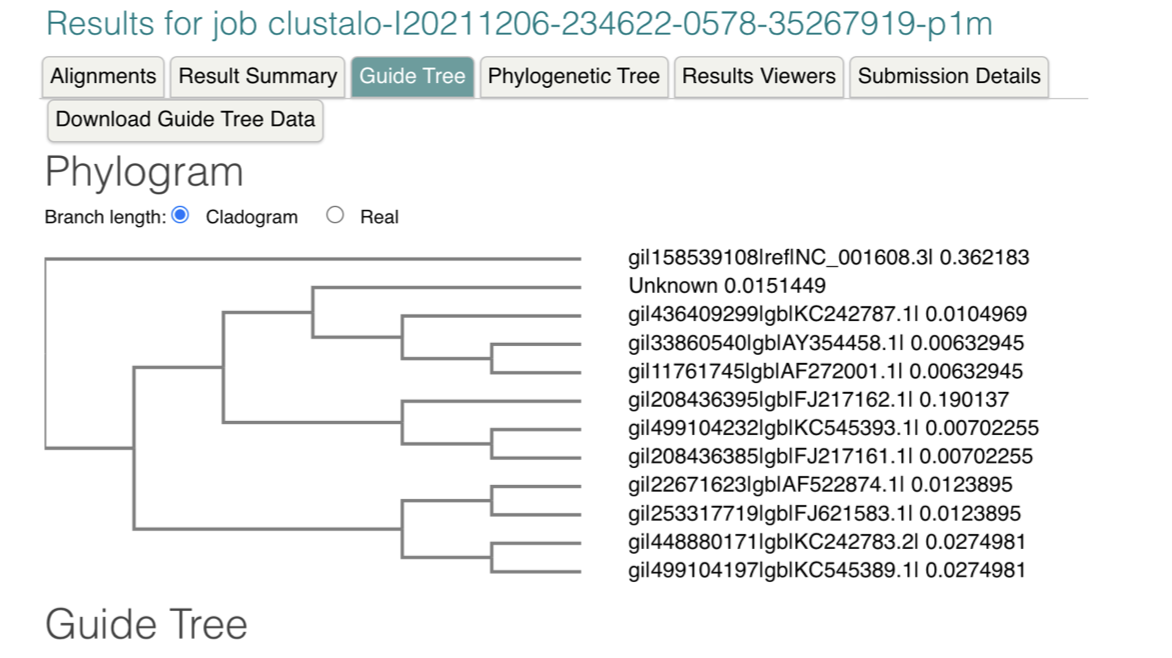
Solved Table 2: NCBI Ebola Virus Genome and Taxonomy

PDF) Group 16SrV phytoplasma in diseased alder trees (Alnus glutinosa) in Lithuania

Cryo-electron Microscopy of Adeno-associated Virus
Recomendado para você
-
 sugarcaramelputty blob but human by prim0078 on DeviantArt04 fevereiro 2025
sugarcaramelputty blob but human by prim0078 on DeviantArt04 fevereiro 2025 -
 New Life Academy - Team Home New Life Academy Eagles Sports04 fevereiro 2025
New Life Academy - Team Home New Life Academy Eagles Sports04 fevereiro 2025 -
 Onassis Airtimes: The event tip 2020 - - Elferspot Magazine04 fevereiro 2025
Onassis Airtimes: The event tip 2020 - - Elferspot Magazine04 fevereiro 2025 -
 marina by prim0078 on DeviantArt04 fevereiro 2025
marina by prim0078 on DeviantArt04 fevereiro 2025 -
 ArtStation - The Detective04 fevereiro 2025
ArtStation - The Detective04 fevereiro 2025 -
 SCP 3934 For Sale Loch Ness Monsters Marshall, Carter & Dark SCP/SCP Pet04 fevereiro 2025
SCP 3934 For Sale Loch Ness Monsters Marshall, Carter & Dark SCP/SCP Pet04 fevereiro 2025 -
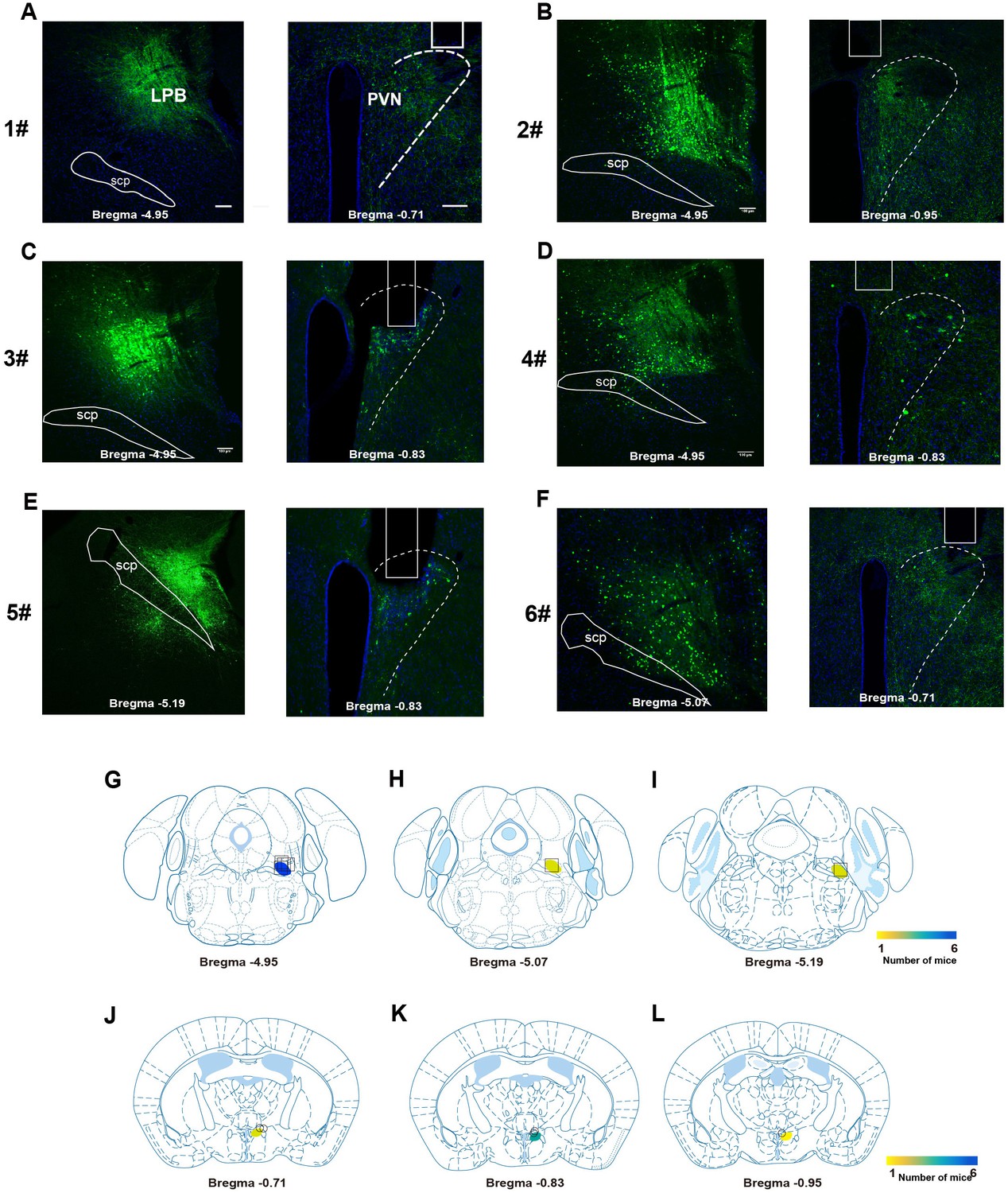 A parabrachial to hypothalamic pathway mediates defensive behavior04 fevereiro 2025
A parabrachial to hypothalamic pathway mediates defensive behavior04 fevereiro 2025 -
 Gelation Mechanism of Poly(N-isopropylacrylamide)−Clay Nanocomposite Gels04 fevereiro 2025
Gelation Mechanism of Poly(N-isopropylacrylamide)−Clay Nanocomposite Gels04 fevereiro 2025 -
 The User Collection - Collection04 fevereiro 2025
The User Collection - Collection04 fevereiro 2025 -
 Who We Are04 fevereiro 2025
Who We Are04 fevereiro 2025
você pode gostar
-
 Anime Expo on X: HIDIVE presents the dub premiere of The Eminence in Shadow – Season 2, Episode 1 w/ special guests Adam Gibbs and Christina Kelly! / X04 fevereiro 2025
Anime Expo on X: HIDIVE presents the dub premiere of The Eminence in Shadow – Season 2, Episode 1 w/ special guests Adam Gibbs and Christina Kelly! / X04 fevereiro 2025 -
Cat Condo 2 for Android - Download the APK from Uptodown04 fevereiro 2025
-
 Chapter 108: Journey's End, Fullmetal Alchemist Wiki04 fevereiro 2025
Chapter 108: Journey's End, Fullmetal Alchemist Wiki04 fevereiro 2025 -
 Listen to One Piece Opening 5 Full Version by Johny Ang in All one piece openings (japanese) playlist online for free on SoundCloud04 fevereiro 2025
Listen to One Piece Opening 5 Full Version by Johny Ang in All one piece openings (japanese) playlist online for free on SoundCloud04 fevereiro 2025 -
 Países Baixos vencem Geórgia em jogo de preparação - CNN Portugal04 fevereiro 2025
Países Baixos vencem Geórgia em jogo de preparação - CNN Portugal04 fevereiro 2025 -
 Ghost (10/10) Movie CLIP - Carl's End (1990) HD04 fevereiro 2025
Ghost (10/10) Movie CLIP - Carl's End (1990) HD04 fevereiro 2025 -
 All photos about Tomodachi Game page 119 - Mangago04 fevereiro 2025
All photos about Tomodachi Game page 119 - Mangago04 fevereiro 2025 -
 Xbox 360 Fat Replacement NO HDMI Console Only - No Cables or Accessories (Renewed)04 fevereiro 2025
Xbox 360 Fat Replacement NO HDMI Console Only - No Cables or Accessories (Renewed)04 fevereiro 2025 -
 Os golpes mais artísticos de Street Fighter V04 fevereiro 2025
Os golpes mais artísticos de Street Fighter V04 fevereiro 2025 -
 animes - Na Nossa Estante04 fevereiro 2025
animes - Na Nossa Estante04 fevereiro 2025