Identifying common transcriptome signatures of cancer by interpreting deep learning models, Genome Biology
Por um escritor misterioso
Last updated 21 março 2025
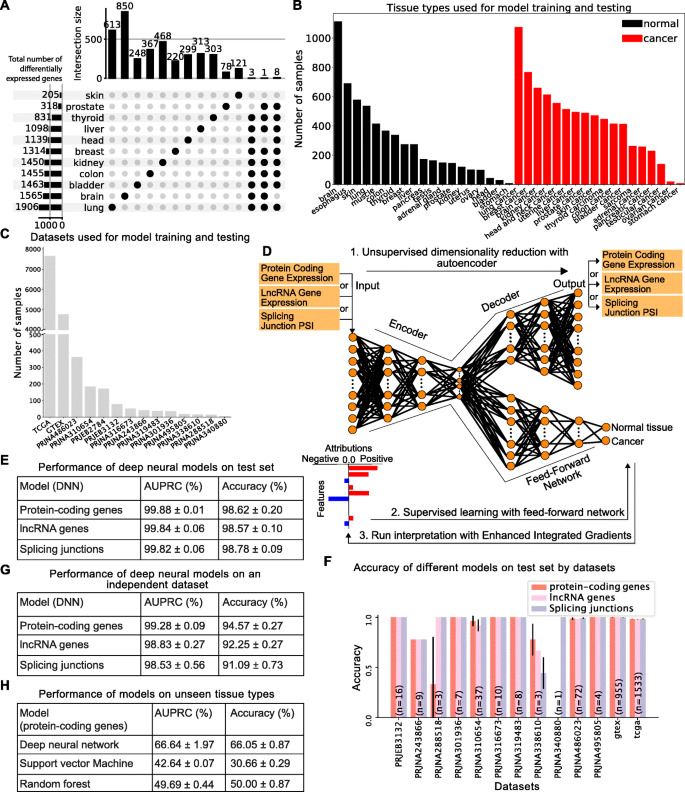
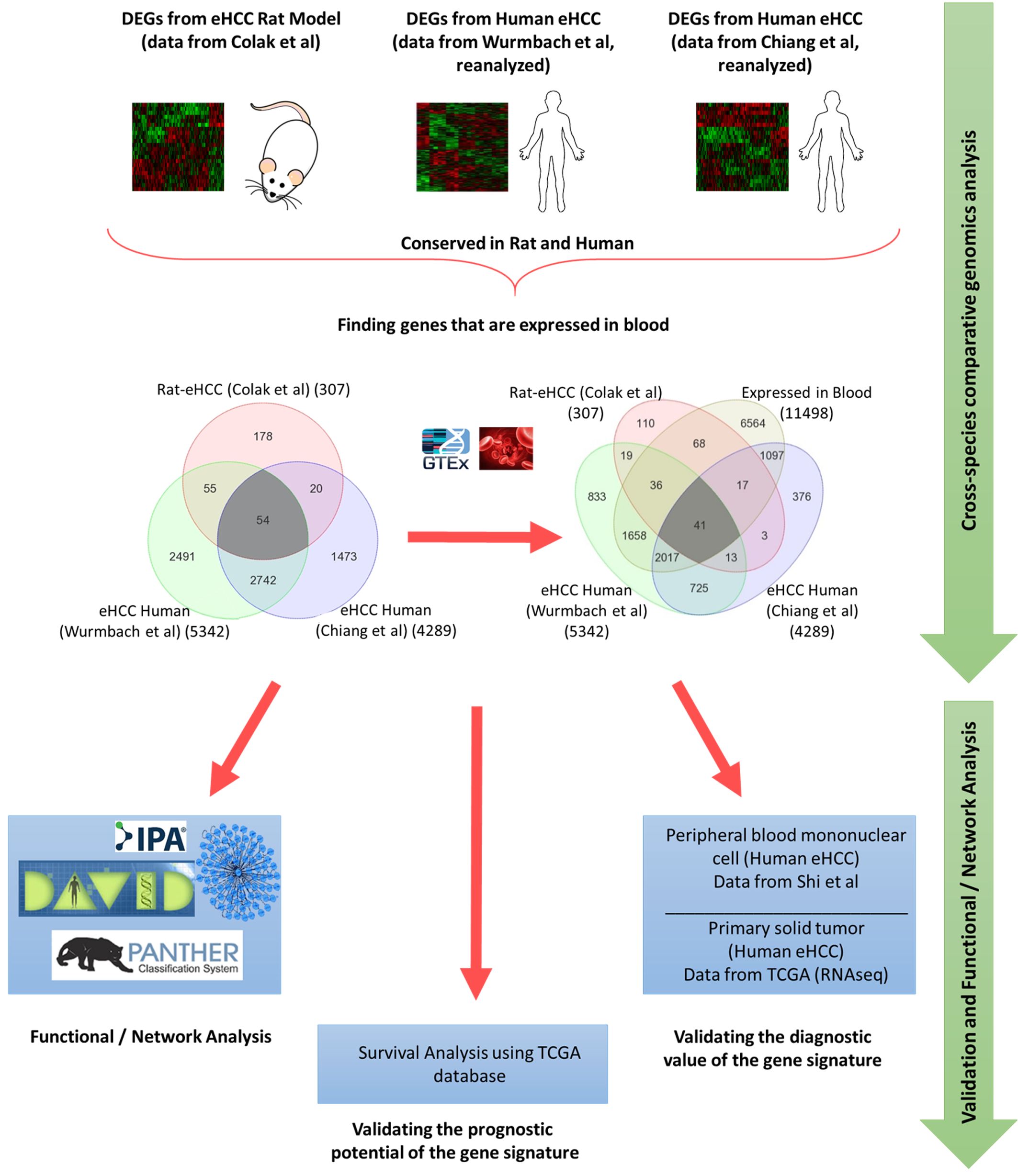
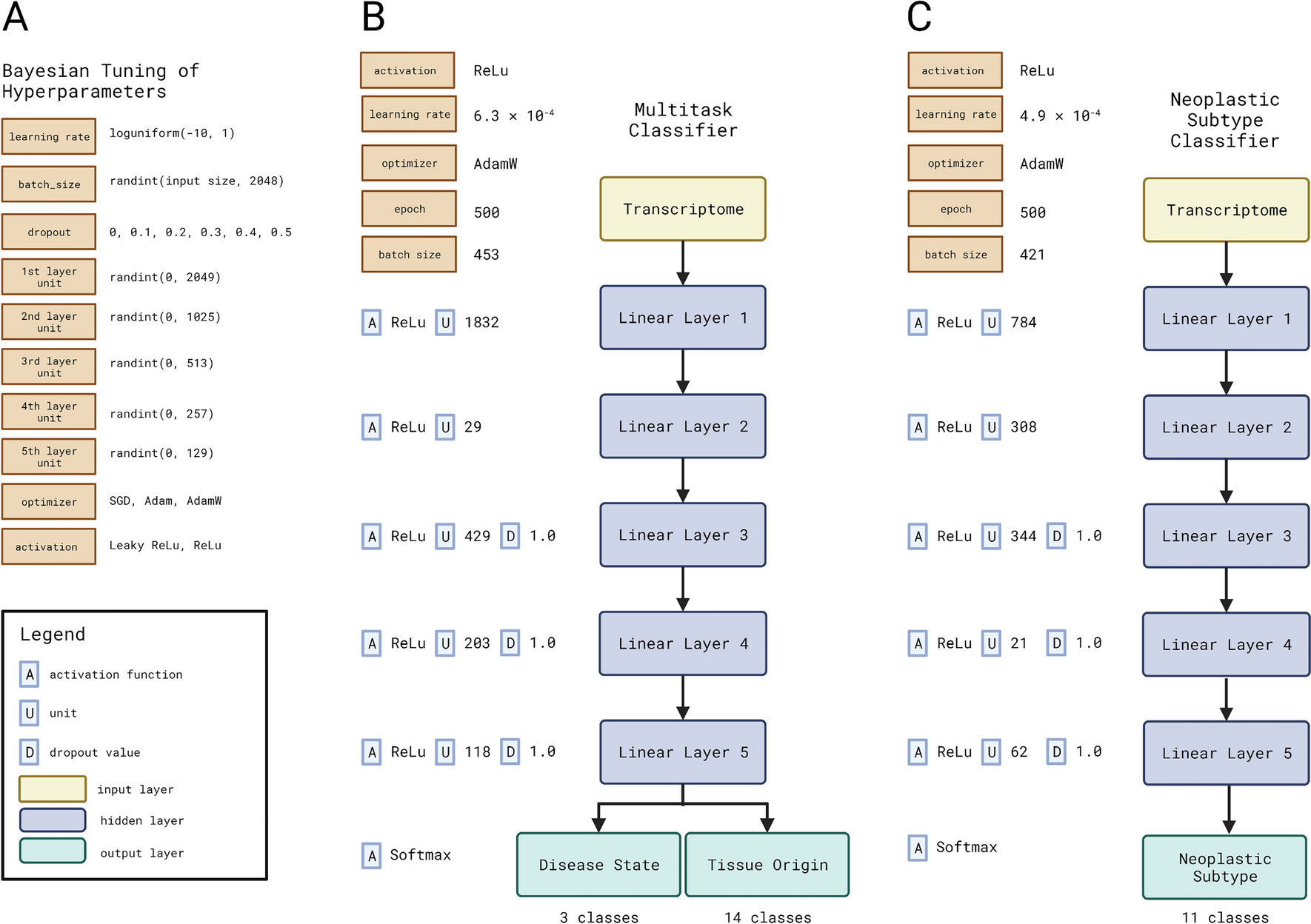
Background Cancer is a set of diseases characterized by unchecked cell proliferation and invasion of surrounding tissues. The many genes that have been genetically associated with cancer or shown to directly contribute to oncogenesis vary widely between tumor types, but common gene signatures that relate to core cancer pathways have also been identified. It is not clear, however, whether there exist additional sets of genes or transcriptomic features that are less well known in cancer biology but that are also commonly deregulated across several cancer types. Results Here, we agnostically identify transcriptomic features that are commonly shared between cancer types using 13,461 RNA-seq samples from 19 normal tissue types and 18 solid tumor types to train three feed-forward neural networks, based either on protein-coding gene expression, lncRNA expression, or splice junction use, to distinguish between normal and tumor samples. All three models recognize transcriptome signatures that are consistent across tumors. Analysis of attribution values extracted from our models reveals that genes that are commonly altered in cancer by expression or splicing variations are under strong evolutionary and selective constraints. Importantly, we find that genes composing our cancer transcriptome signatures are not frequently affected by mutations or genomic alterations and that their functions differ widely from the genes genetically associated with cancer. Conclusions Our results highlighted that deregulation of RNA-processing genes and aberrant splicing are pervasive features on which core cancer pathways might converge across a large array of solid tumor types.

A Comparative Analysis of Single-Cell Transcriptome Identifies
Frontiers Identification of Gene Signature as Diagnostic and

Machine learning identifies signatures of macrophage reactivity
PDF) Identifying common transcriptome signatures of cancer by
A deep learning model to classify neoplastic state and tissue
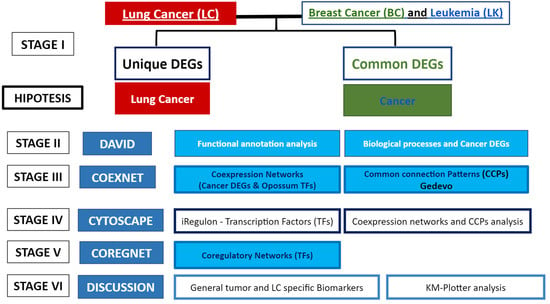
Biology, Free Full-Text
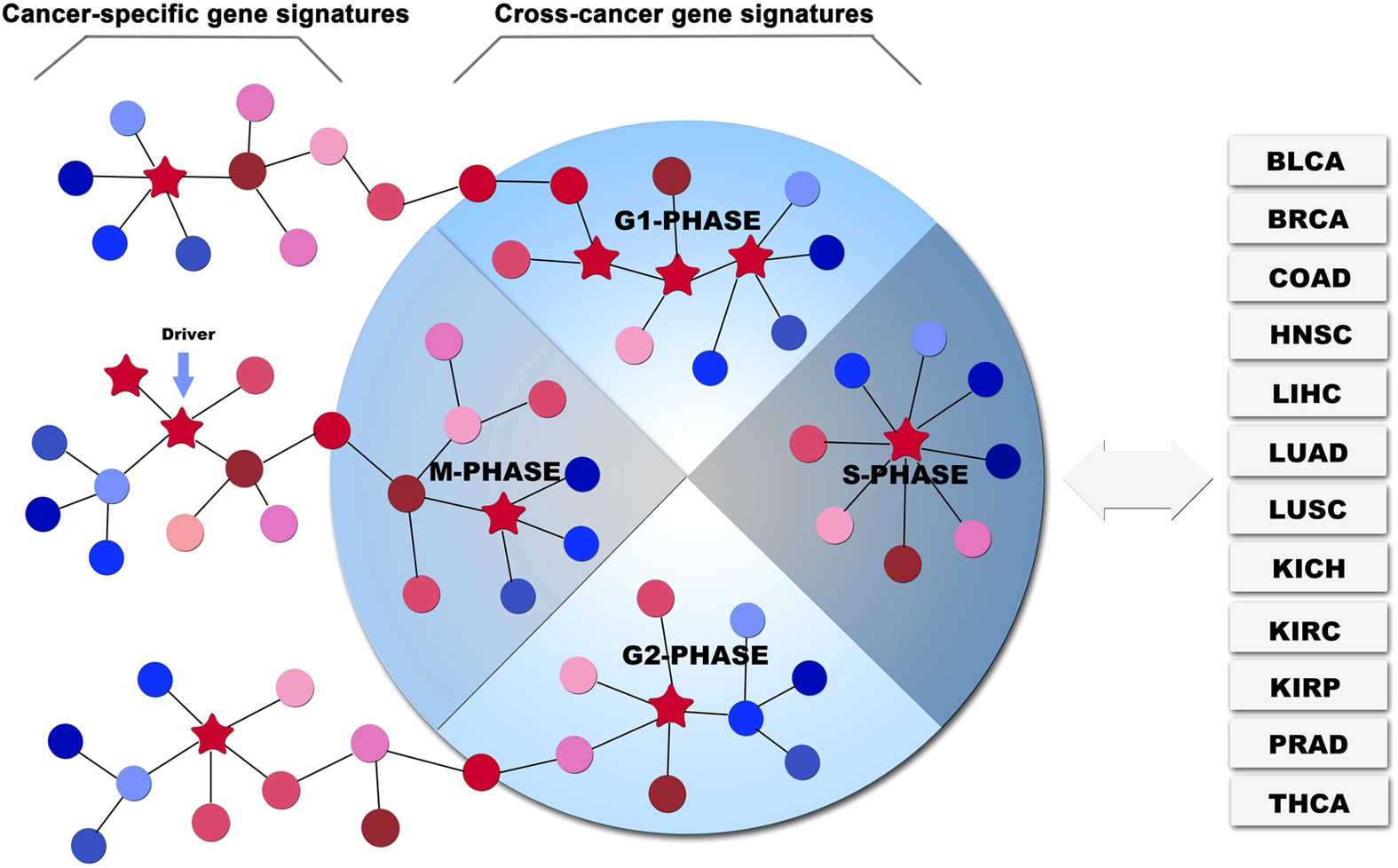
Large-scale RNA-Seq Transcriptome Analysis of 4043 Cancers and 548
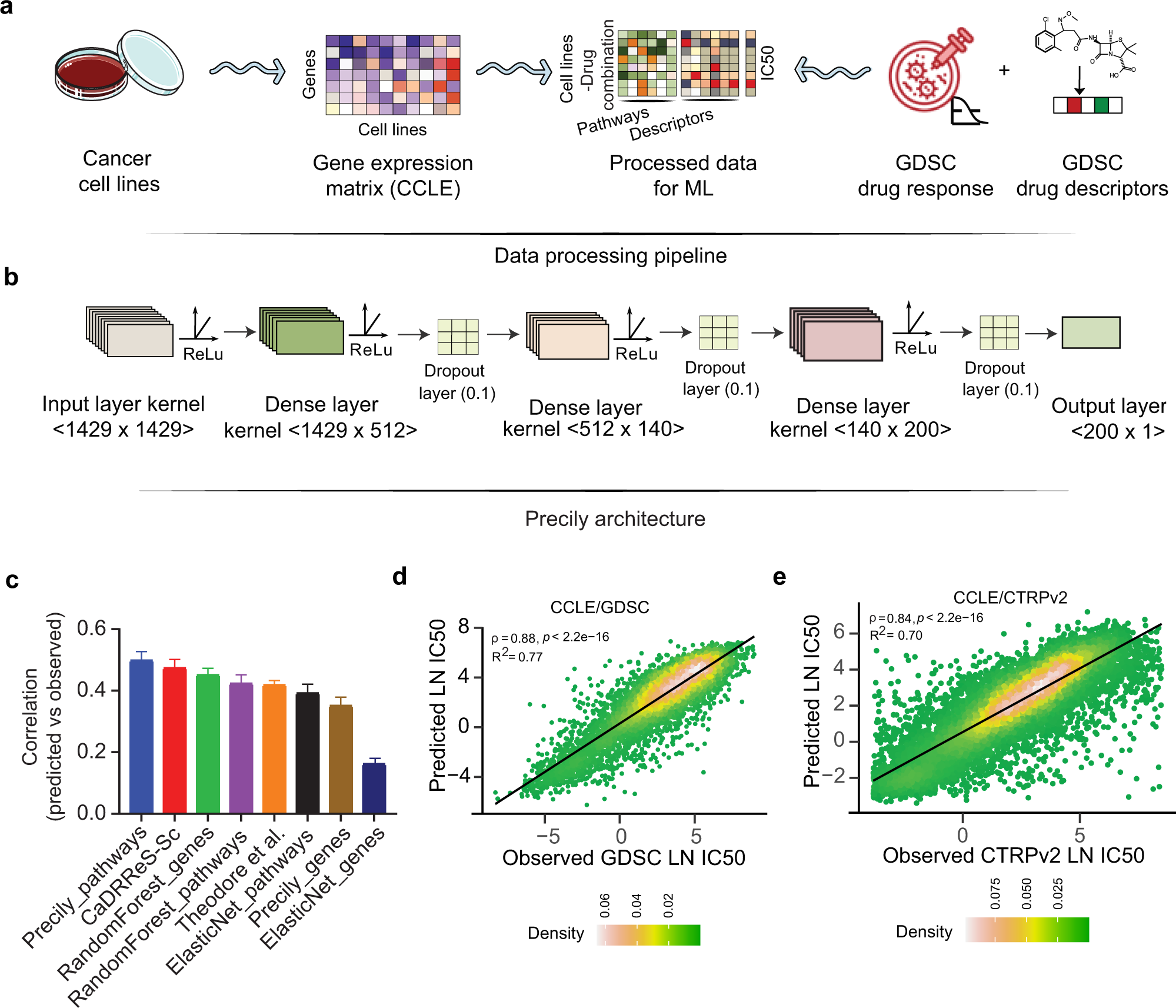
Gene expression based inference of cancer drug sensitivity
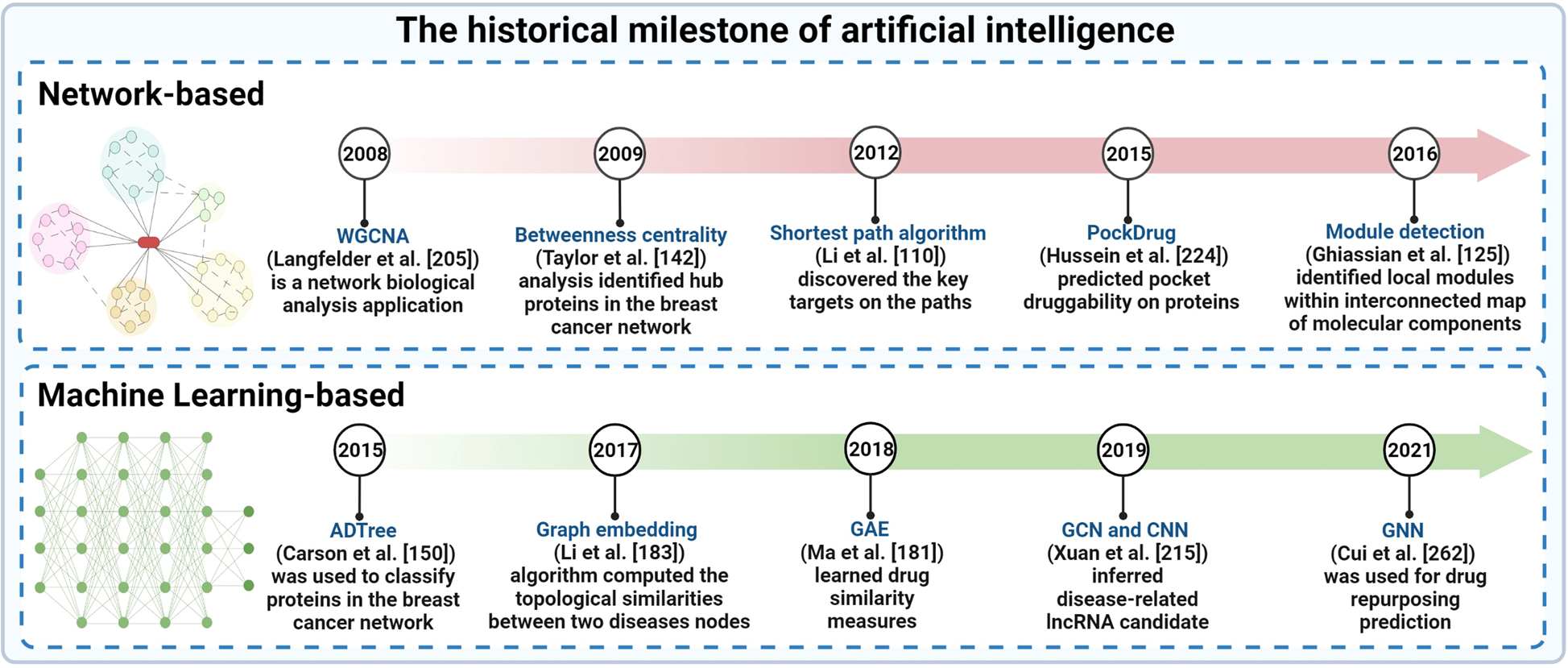
Artificial intelligence in cancer target identification and drug
Recomendado para você
-
 Brain Test Level 367 It's cold the fireplace needs more fire in 202321 março 2025
Brain Test Level 367 It's cold the fireplace needs more fire in 202321 março 2025 -
 Brain Test Level 372 He wants big muscles in 202321 março 2025
Brain Test Level 372 He wants big muscles in 202321 março 2025 -
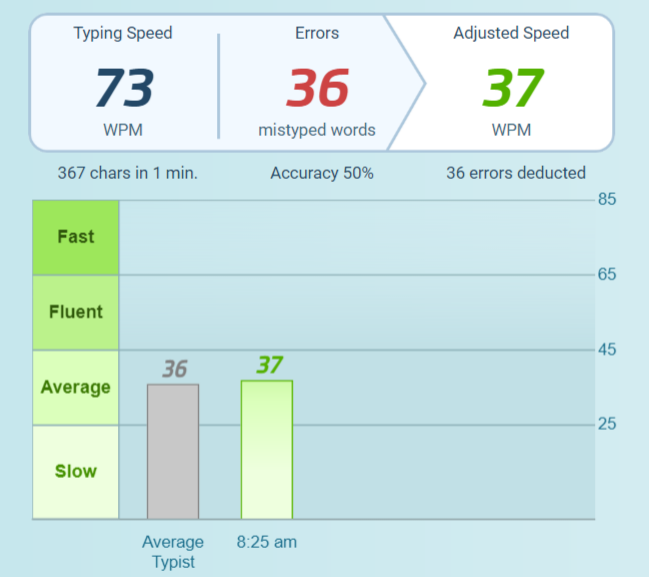 Tech Thursday: Voice-to-Text - by Breana Bayraktar21 março 2025
Tech Thursday: Voice-to-Text - by Breana Bayraktar21 março 2025 -
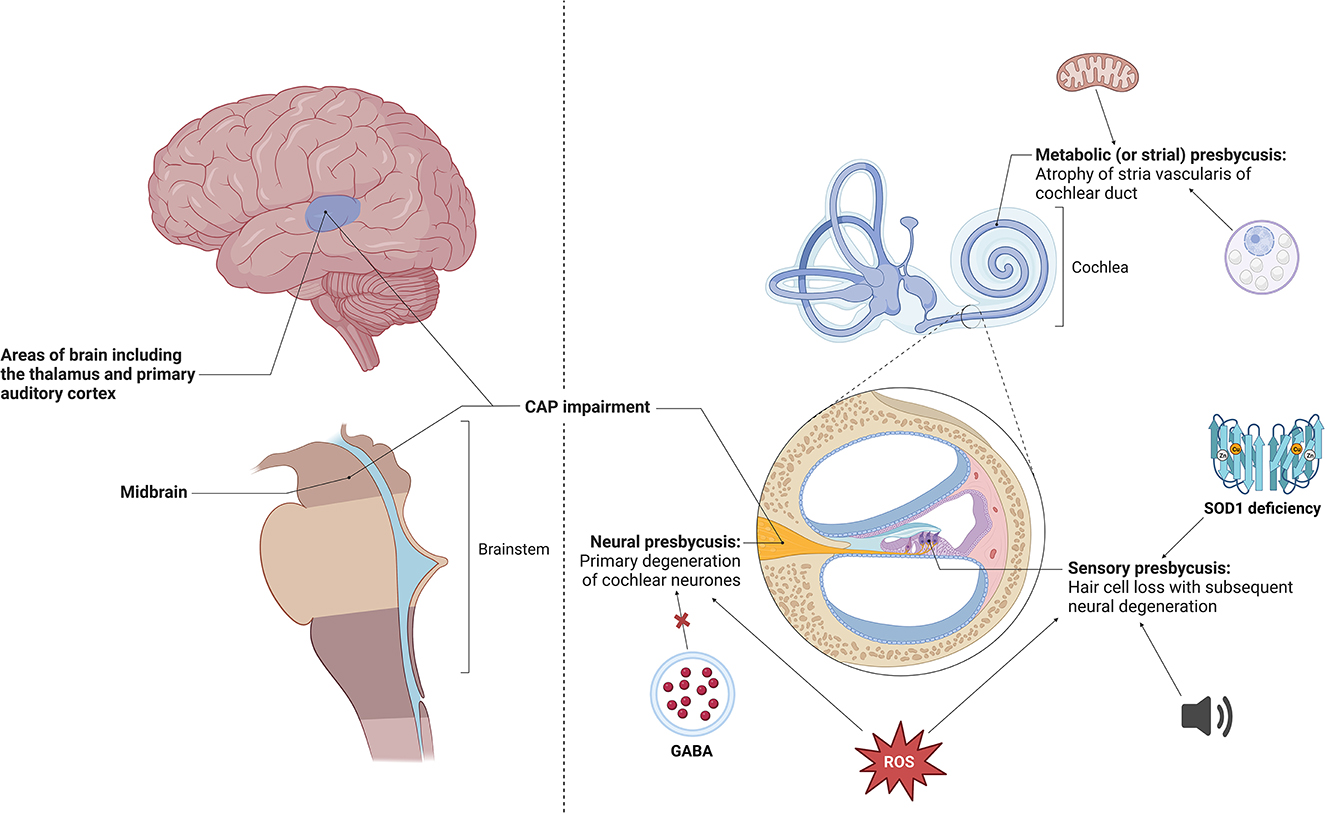 Frontiers Hearing loss and its link to cognitive impairment and dementia21 março 2025
Frontiers Hearing loss and its link to cognitive impairment and dementia21 março 2025 -
 Recent advancements in understanding fin regeneration in zebrafish - Sehring - 2020 - WIREs Developmental Biology - Wiley Online Library21 março 2025
Recent advancements in understanding fin regeneration in zebrafish - Sehring - 2020 - WIREs Developmental Biology - Wiley Online Library21 março 2025 -
 Ryuta Kawashima: The devil who cracked the dementia code, The Independent21 março 2025
Ryuta Kawashima: The devil who cracked the dementia code, The Independent21 março 2025 -
 Blinking a Book - Create Your Own Comic Strips Online with MakeBeliefsComix21 março 2025
Blinking a Book - Create Your Own Comic Strips Online with MakeBeliefsComix21 março 2025 -
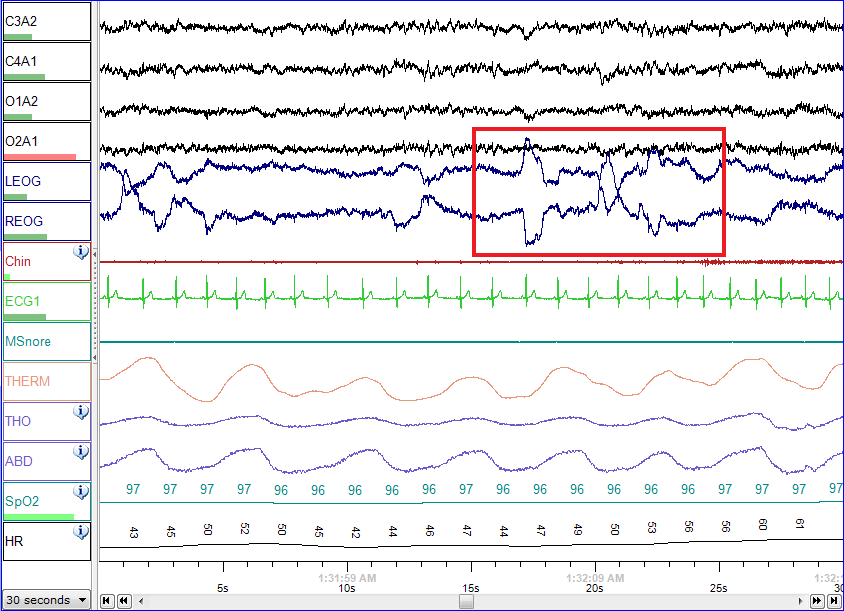 Polysomnography - Wikipedia21 março 2025
Polysomnography - Wikipedia21 março 2025 -
 Fluid transport in the brain21 março 2025
Fluid transport in the brain21 março 2025 -
Men's Health of Parkersburg21 março 2025
você pode gostar
-
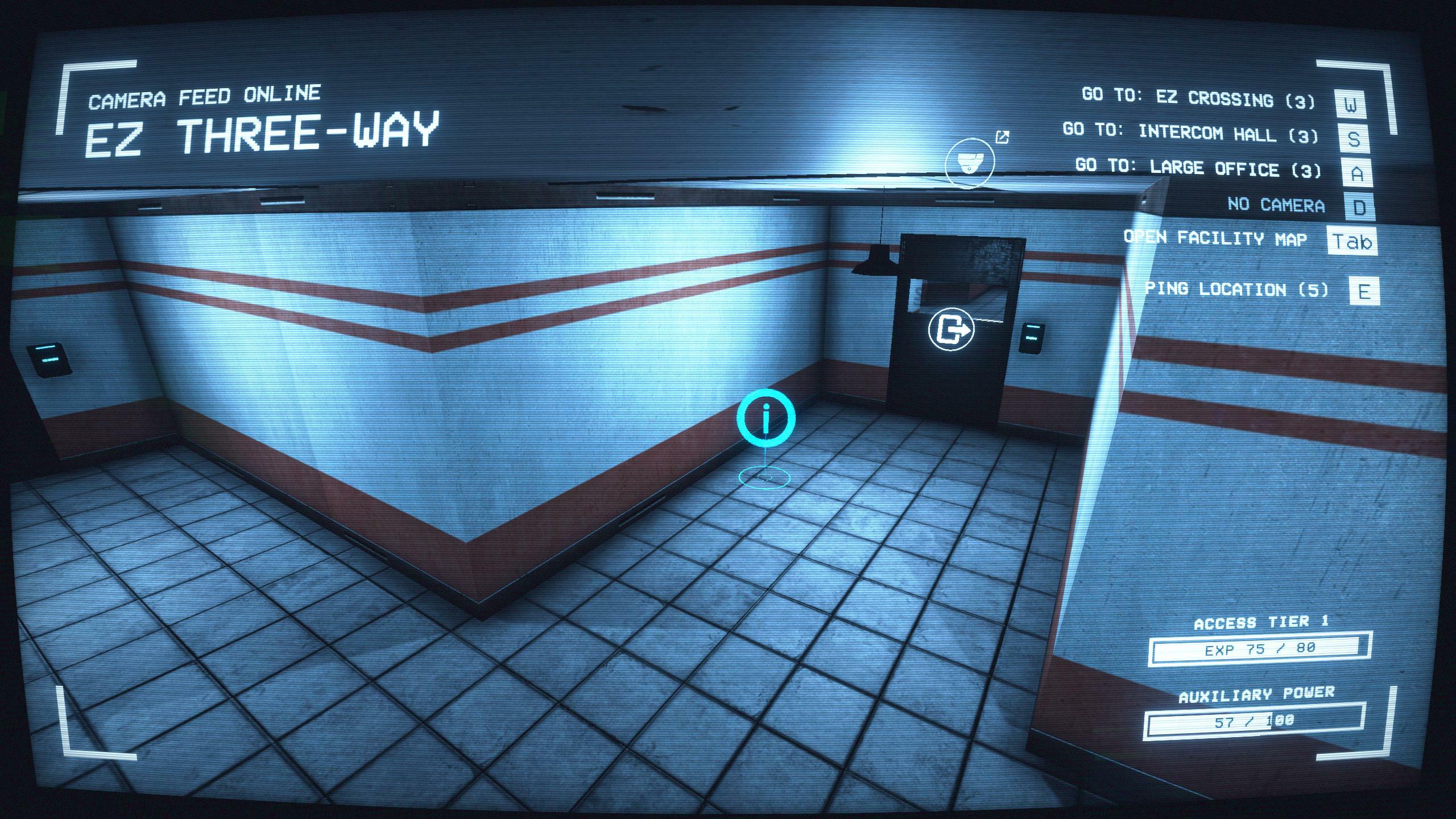 079 Soft Rework · SCP: Secret Laboratory update for 24 August 202221 março 2025
079 Soft Rework · SCP: Secret Laboratory update for 24 August 202221 março 2025 -
 Harry Potter: Xadrez de Bruxo!21 março 2025
Harry Potter: Xadrez de Bruxo!21 março 2025 -
 365 Caça Palavras Cruzadas Médio Passatempo Livro Culturama21 março 2025
365 Caça Palavras Cruzadas Médio Passatempo Livro Culturama21 março 2025 -
![A few memes from my collection [669] — Steemit](https://steemitimages.com/640x0/https://cdn.steemitimages.com/DQmTjgREpJTLdgZd3Q3nA885eaXdMNGu7nBvhm8KdFWFwPg/1659956493151_result_result.jpg) A few memes from my collection [669] — Steemit21 março 2025
A few memes from my collection [669] — Steemit21 março 2025 -
 FREE! - Simple Flower Page Border, Page Borders21 março 2025
FREE! - Simple Flower Page Border, Page Borders21 março 2025 -
 Dinossauros Para Colorir Lion coloring pages, Dinosaur coloring pages, Dinosaur coloring21 março 2025
Dinossauros Para Colorir Lion coloring pages, Dinosaur coloring pages, Dinosaur coloring21 março 2025 -
 WARNER BROS GAMES - Hogwarts Legacy for Sony Playstation PS521 março 2025
WARNER BROS GAMES - Hogwarts Legacy for Sony Playstation PS521 março 2025 -
 Prime Video21 março 2025
Prime Video21 março 2025 -
 Driver Booster PRO Review - Keep Your Device Drivers and Game Components Updated21 março 2025
Driver Booster PRO Review - Keep Your Device Drivers and Game Components Updated21 março 2025 -
Bola de basquete silenciosa da @Hoop City 🇧🇷 #basketball #basquete , silent basketball21 março 2025

